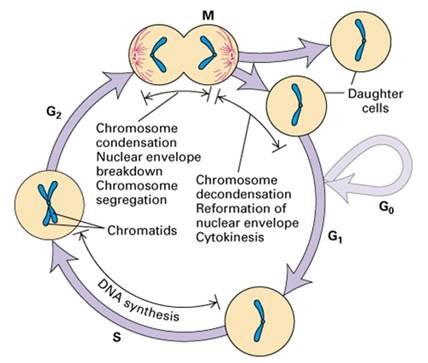
Principles of flow cytometry analysis of the cell cycle
In different phases of the cell cycle, there are differences in DNA content, staining with DNA dyes (such as the most widely used dye PI), the DNA content is high, and the DNA content is low, so the cell can be judged according to the change in DNA fluorescence intensity.
To analyze the cell cycle using PI, RNase needs to be added to eliminate the effects of RNA
PI stains nucleic acids by inserting bases superimposed in the double helix structure, and it can be inserted into all DNA and RNA double-stranded regions, PI is not DNA-specific, so RNase needs to be added to eliminate the effect of RNA.
When the cell cycle is tested, it must not be washed
Unlike the firm bond between the protein dye and the cell, the binding of DNA to the dye is a loose balance between the bound state and the free state, which is not very strong, so the washing step will cause the dye to be lost, reduce the specific fluorescence of the nucleic acid, and cause the measured value to become smaller, so it is important not to wash the DNA when staining.
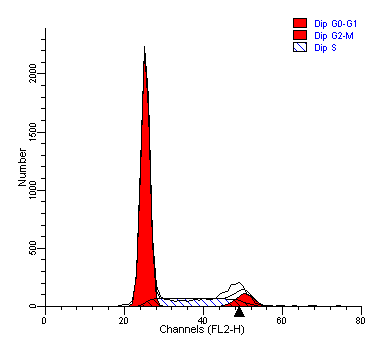
Cell cycle assays for fixed and live cells
Dyes such as PI, 7-AAD, TOTO-3 and YOYO-1 cannot penetrate the intact cell membrane, and during staining, it is generally necessary to fix and permeabilize the cell membrane with 70% ethanol, so that these dyes can enter the nucleus and bind to DNA, so many cell cycle detection kits on the market need to fix cells; If you want to test live cell samples, you can choose LinkBio's cell cycle assay kit, which can detect not only fixed cells, but also live cells.
A Live-Cell Protocol
- Collect cells by centrifugation, wash once, and discard the supernatant
- Add 1 mL of DNA Staining Solution (A) and 10 uL of Permeabilization Solution (B)
- Incubate for 30 minutes at room temperature in the dark
- Flow cytometry assay
B Fixed cell protocol
- Carefully fix cells and store at -20°C
- Collect cells by centrifugation, wash once, and discard the supernatant
- Add 1mL of DNA Staining Solution (A)
- Incubate for 30 minutes at room temperature in the dark
- Flow cytometry assay
Other issues that should be noted
• Instrument: Regular cleaning of the nozzle, sufficient laser warm-up time, low flow rate injection, etc.
• Sample: Digestion should be sufficient, but not excessive, to ensure that the cells are not adhered or broken, and the staining should be sufficient.

